IDO1 and IDO2 catalyze the first step in the kynurenine pathway: The conversion of tryptophan to N-formyl-kynurenine. N-formyl-kynurenine can be converted to kynurenine (KYN). KYN can be further processed to kynurenic acid (KYNA).
The ATPase Inhibitory Factor (ATPIF1) is a master regulator of energy metabolism and of cell survival. (4).
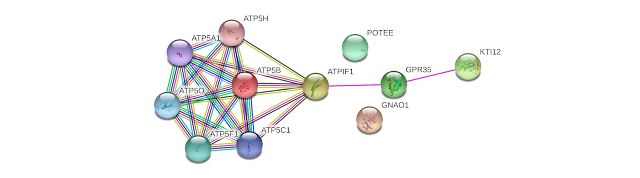
The kynurenic acid responsive GPR35 interacts with ATPIF1. And ATPIF1 interacts with ATP5B (5).
ATP synthase subunit beta expression was upregulated in ME patients (7).
Figure from Genecards STRING interaction network (5).
GPR35: G-protein coupled receptor 35; Acts as a receptor for kynurenic acid, an intermediate in the tryptophan metabolic pathway. The activity of this receptor is mediated by G-proteins that elicit calcium mobilization and inositol phosphate production through G(qi/o) proteins (5).
ATPIF1: ATPase inhibitor, mitochondrial; Endogenous F(1)F(o)-ATPase inhibitor limiting ATP depletion when the mitochondrial membrane potential falls below a threshold and the F(1)F(o)-ATP synthase starts hydrolyzing ATP to pump protons out of the mitochondrial matrix. Required to avoid the consumption of cellular ATP when the F(1)F(o)-ATP synthase enzyme acts as an ATP hydrolase. Indirectly acts as a regulator of heme synthesis in erythroid tissues- regulates heme synthesis by modulating the mitochondrial pH and redox potential (5).
ATP5B: ATP synthase subunit beta, mitochondrial; Mitochondrial membrane ATP synthase (F(1)F(0) ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain. F-type ATPases consist of two structural domains, F(1) - containing the extramembraneous catalytic core, and F(0) - containing the membrane proton channel, linked together by a central stalk and a peripheral stalk (5).
Is something going on in the ATP5B - ATPIF1 - GPR35 interaction network in ME?
Further reading:
Is the kynurenic acid responsive Gpr35 involved in the ME pathomechanism?http://followmeindenmark.blogspot.com/2019/06/is-kynurenic-acid-responsive-gpr35.html
Complex V is down in ME - does it also explain Electromagnetic Hypersensitivity?
http://followmeindenmark.blogspot.com/2019/07/complex-v-is-down-in-me-does-it-also.html
Complex V is down in ME - does it explain Chemical Intolerance?
http://followmeindenmark.blogspot.com/2019/07/complex-v-is-down-in-me-does-it-explain.html
Mutations in the IDO2 gene and DNA methylations in genes in the NAD/NADP synthesis pathway in ME
http://followmeindenmark.blogspot.com/2019/07/mutations-in-ido2-gene-and-dna.html
CTLA-4 induces IDO and SOCS3 drives degradation of IDO
http://followmeindenmark.blogspot.com/2019/06/ctla-4-induces-ido-and-socs3-drives.html
References:
1) Kashi AA Davis RW and, Phair RD: The IDO Metabolic Trap Hypothesis for the Etiology of ME/CFS. Diagnostics (Basel). 2019 Jul 26;9(3). pii: E82. doi: 10.3390/diagnostics9030082. https://www.mdpi.com/2075-4418/9/3/82
Metabolic Traps in ME/CFS - Research Update by Dr. Robert Phair
https://www.youtube.com/watch?v=Quh-77gvw4Q
2) Missailidis, D.; Annesley, S.J.; Allan, C.Y.; Sanislav, O.; Lidbury, B.A.; Lewis, D.P.; Fisher, P.R. An isolated Complex V defect and dysregulated mitochondrial function in immortalized lymphocytes from ME/CFS patients. Submitted 2019.
Specific Mitochondrial Respiratory Defects & Compensatory Changes in ME/CFS Patient Cells
https://www.youtube.com/watch?v=SjK39QCPeeY
3) Wikipedia: ATP synthase: https://en.wikipedia.org/wiki/ATP_synthase
4) García-Bermúdez J, Cuezva JM.:The ATPase Inhibitory Factor (IF1) is a master regulator of energy metabolism and of cell survival. Biochim Biophys Acta. 2016 Aug;1857(8):1167-1182. doi: 10.1016/j.bbabio.2016.02.004. Epub 2016 Feb 12.
https://www.sciencedirect.com/science/article/pii/S0005272816300238?via%3Dihub#f0020
5) Genecards STRING interaction network, GPR35 https://www.genecards.org/
6) Trivedi et al: Identification of ME/CFS - associated DNA methylation patterns.
Plos One 2018, 13, 7 https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0201066
7) Ciregia et al: Bottom-up proteomics suggests an association between differential expression of mitochondrial proteins and chronic fatigue syndrome. Transl Psychiatry. 2016 Sep 27;6(9):e904. doi: 10.1038/tp.2016.184. https://www.nature.com/articles/tp2016184
6) Trivedi et al: Identification of ME/CFS - associated DNA methylation patterns.
Plos One 2018, 13, 7 https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0201066
7) Ciregia et al: Bottom-up proteomics suggests an association between differential expression of mitochondrial proteins and chronic fatigue syndrome. Transl Psychiatry. 2016 Sep 27;6(9):e904. doi: 10.1038/tp.2016.184. https://www.nature.com/articles/tp2016184
Ingen kommentarer:
Send en kommentar
Bemærk! Kun medlemmer af denne blog kan sende kommentarer.